Visualization Community
Explore, create, and analyze
genomic and epigenomic visualizations.
Contributing Organizations

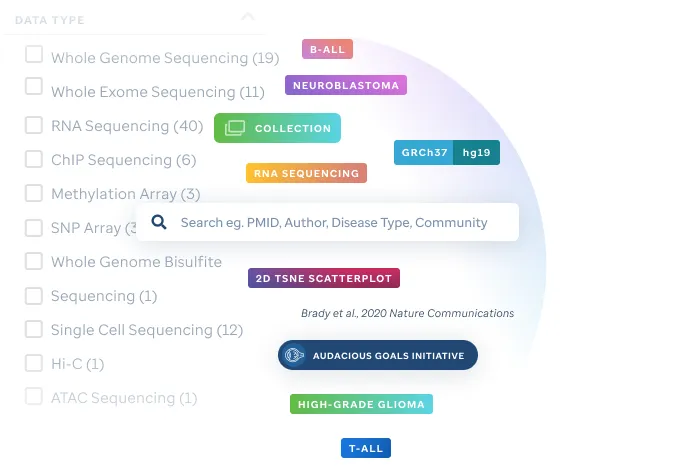
Search through our growing number of publication quality visualizations.
Easily identify genomic data visualizations associated with select pediatric cancers and other diseases via filtering on publication, project, institution or diagnosis.
Explore Visualizations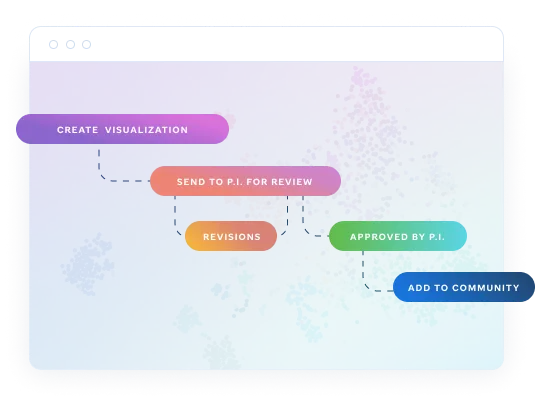
Create, review, and make your research public.
Any visualization you or your team create is private by default. This ensures privacy - P.I. approval is required to make visualizations publicly viewable once completed or published.

Build with the best.
Our state of the art tools render robust visualizations for quick analysis of simple or complex datasets.
GenomePaint
Interactive visualization platform for integrating whole-genome, whole-exome, transcriptome, and epigenomic data of tumor samples.
ProteinPaint
Visualize over 22 tracks to compare any set of variants in a gene in the context of protein domains and other information.

Allelic imbalance (aicheck)
Displays the allelic imbalance of the heterozygous SNP markers in a tumor genome as compared to this patient’s germline genome, as a way of indicating loss-of-heterozygosity.

Arc (hicstraw)
Used to display chromatin capture data by focusing on the frequency of links between distant genomic regions.

BAM
Visualizes the aligned reads and assists in understanding depth and quality of genomically mapped features on a nucleotide scale.

BigWig
This format shows the depths of reads in RNA seq, ChIP seq, or genomic sequence as aligned to a reference genome.

JSON BED (bedj)
This customizable bed format shows defined genomic regions identified by ChIP seq, mass spec or other means.

Lollipop plot
Used for displaying the frequency and type of mutation for a given gene on the protein structure in the context of functional domains.

MA+ Volcano plot
The MA plot contrasts the magnitude of expression (M) on the x axis against the amplitude (A) of change between two states. The volcano plot compares the amplitude of change (x-axis ) to the statistical reliability of that change (-log transformed p value, y-axis).

Mutation Landscape (heatmap)
Used for displaying the frequency of various mutations, CNVs and SVs in cancer relevant genes across a cohort of samples from one or many diagnoses.

Splice Junction Cohort (mdsjuction)
This visualization displays the number and color codes the type of mapped junction reads precisely along with a gene model for multiple samples including canonical and non-canonical splice forms.

Stranded BigWig
RNA-seq coverage data as a pair of bigWig tracks, with forward strand on top and reverse strand on bottom.

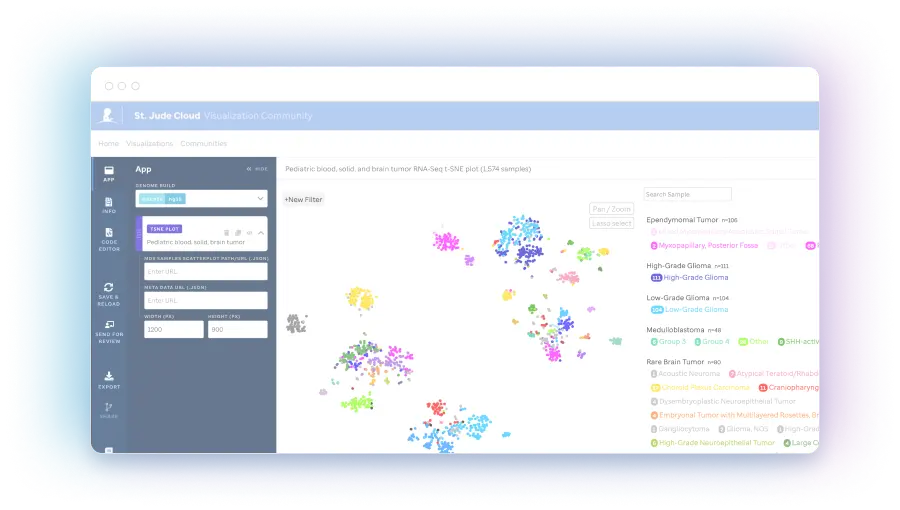
tSNE plot (Scatterplot)
tSNE plots are a display that converts high dimensional data to an interpretable proximity plot such that similarity may be assessed across RNA-seq profiles.

UMAP (single cell)
A data driven dimensional reduction method used to display single cell RNA seq data from a large number of cells.

VCF
This track localizes the variant data on reference genomes and can render frequency data for cohorts.
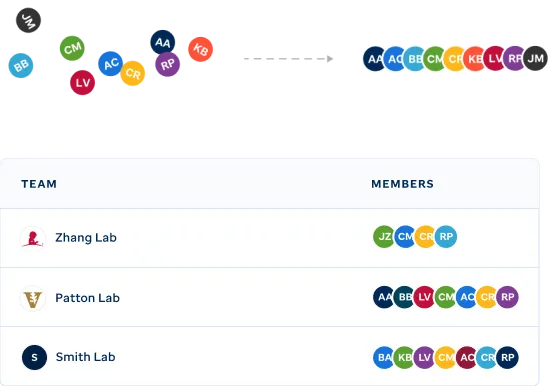
Organize your team's visualizations.
Easily identify genomic data visualizations associated with select pediatric cancers and other diseases via filtering on publication, project, institution or diagnosis.
View your teamsFrom private research to published interactive figures.
Increase the profile of your published visualizations and accessibility of your figures.
Public
Once reviewed by a P.I. or Team lead, a visualization can be made publicly accessible.
Published
Once your research has been published, add your PMID to your visualization to link with the associated publication.
Collections
Organize multiple visualizations into a collection associated with your research.
The expression landscape of 588 RNA-Seq brain tumor samples - of which 459 are fresh frozen and 129 are FFPE samples. These are displayed via a t-SNE plot generated using the St. Jude Cloud RNA-Seq Expression Classification analysis workflow. Diagnosis subtype...
Gene expression profiling of 1,988 B-cell acute lymphoblastic leukemia (B-ALL) cases shown in a two-dimensional tSNE plot (representative of Fig. 1a of Gu et al. 2019) employing the top 1,000 most variable genes. B-ALL subtypes are highlighted in different col...
Selected visualizations related to improving quality of life for pediatric cancer survivors.

Unify teams and labs together in one location.
Easily showcase your work and evolve the community of research.
View Communities

Need more help?
Access our help guides whether it be on how to cite us, who to contact for support, or to find our curated examples with sample data to start generating your own visualizations.
Visit the Docs