NetAD
NetAD: Network modeling of bulk and single-cell multi-omics data to identify drivers for Alzheimer's Disease.
To increase our understanding of Alzheimer's disease (AD) by using network-based systems biology tools to harmonize cross-studied multi-omics data at the bulk and single-cell levels, thereby providing new therapeutic targets and biomarkers towards AD prevention and treatment.
The NetAD community provides accessible data for protein and pathway searches, lightweight data analysis for gene, protein, and phosphopeptide abundance visualization. It presents integrated multi-omics data in a familiar genome browser or proteinPaint display with controlled data access. Additionally, NetAD is a network visualization and analysis platform for existing gene module visualization and protein/gene lookup that enables focal highlighting within the network.
Featured Data:
a) A barplot of protein abundance is displayed when searching a gene or protein name. The barplot summarizes protein abundance across samples (see link here)
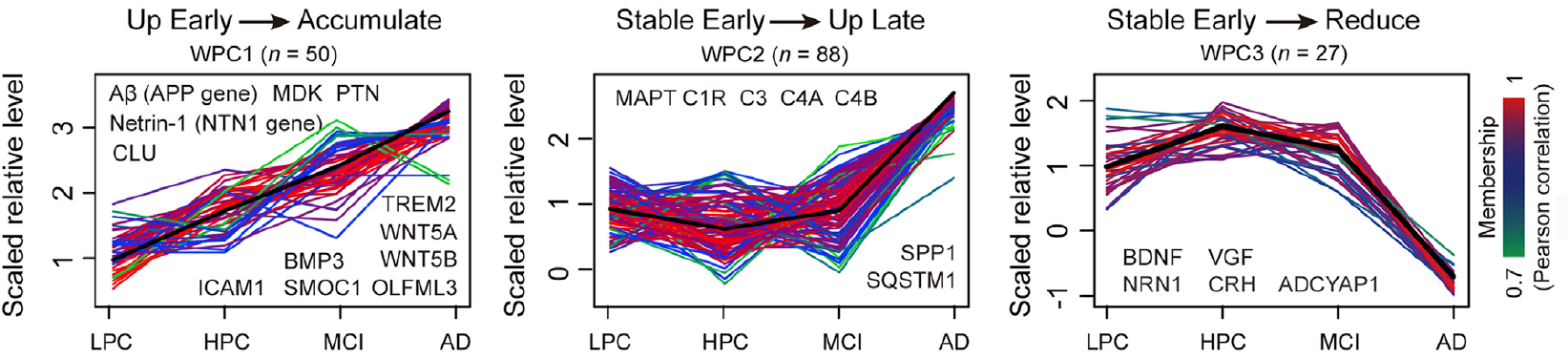
b) Co-expression protein clusters:
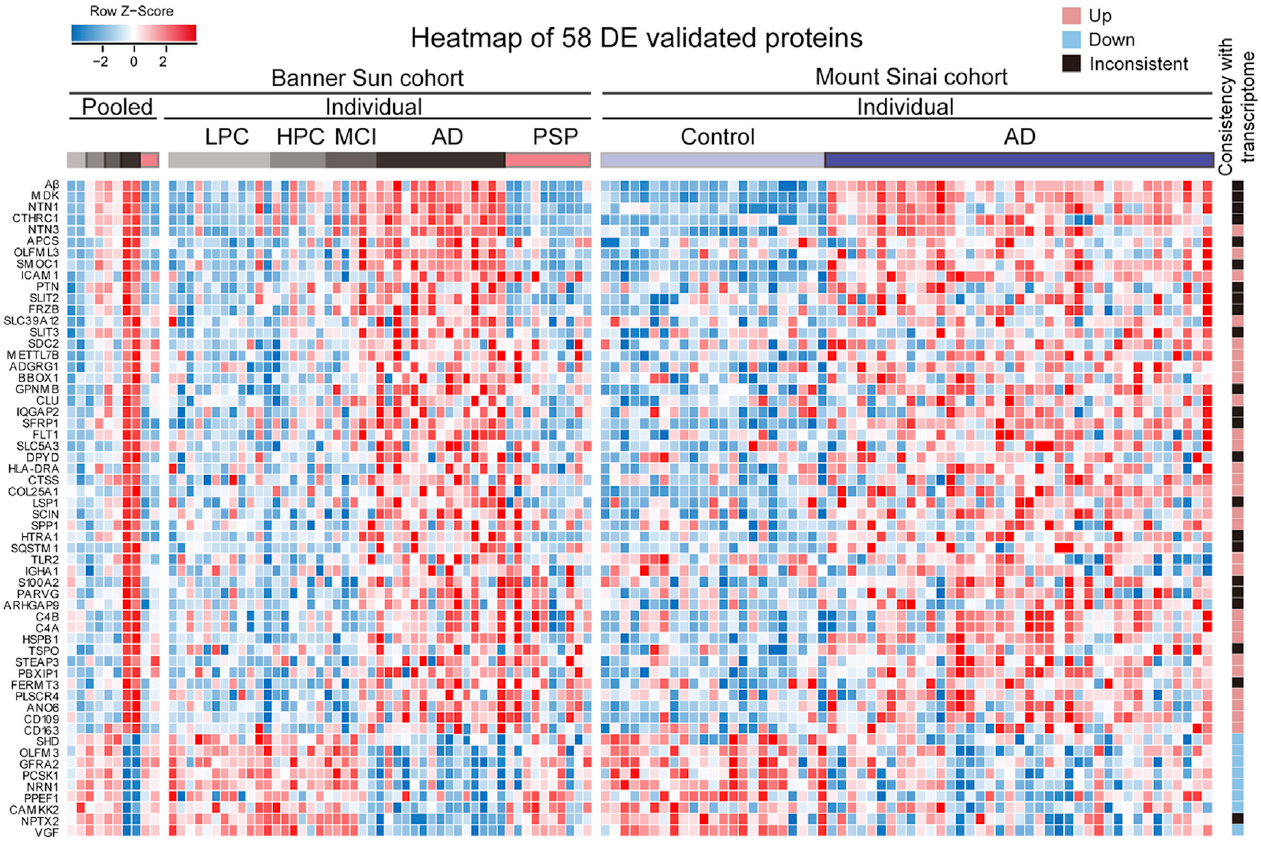
c) A heatmap of normalized protein abundance for proteins within a defined pathway appears when typing a pathway name. The visualized heatmap of proteins within a pathway is displayed across a cohort of samples:
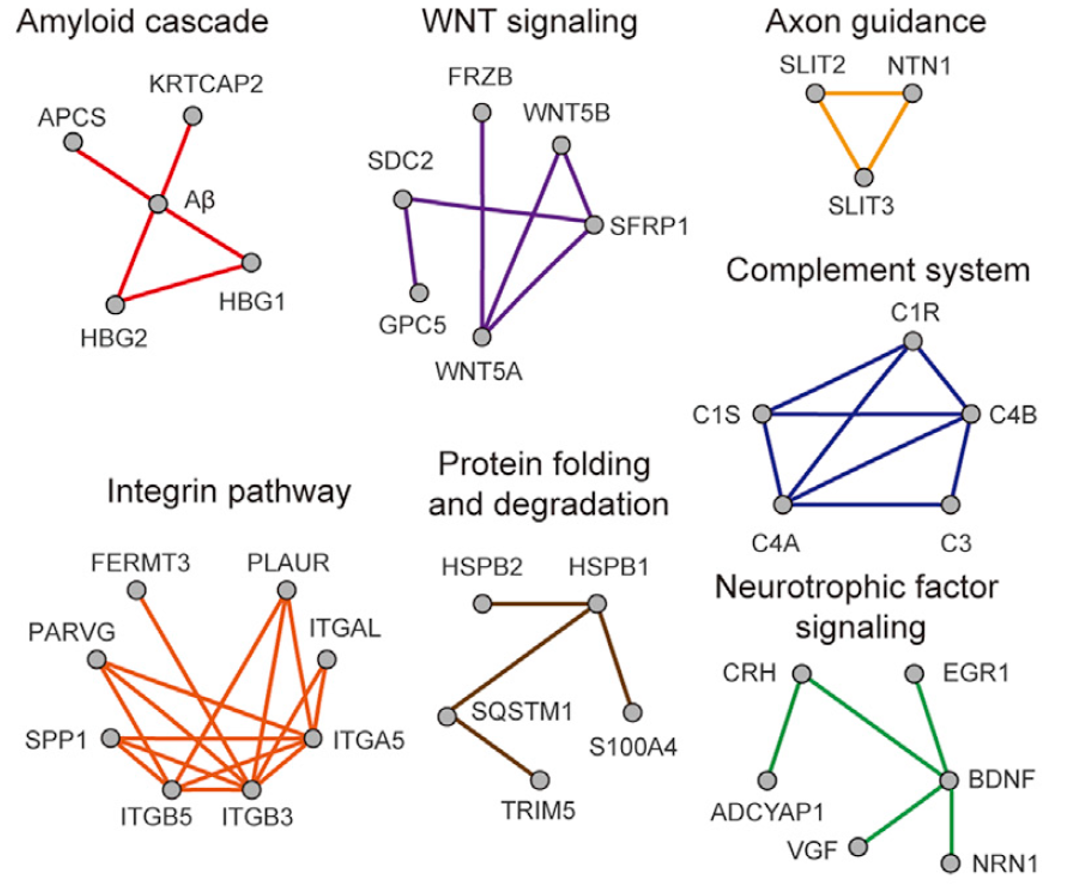
d) A visualized subnetworks of protein-protein interactions:
References
Panels a), b) and f) are from Bai et al., Neuron 2020
Panels c) and d) are from Tan et al., Immunity 2017
Panel e) (Coming soon) is from Li et al., J Proteome Res 2016